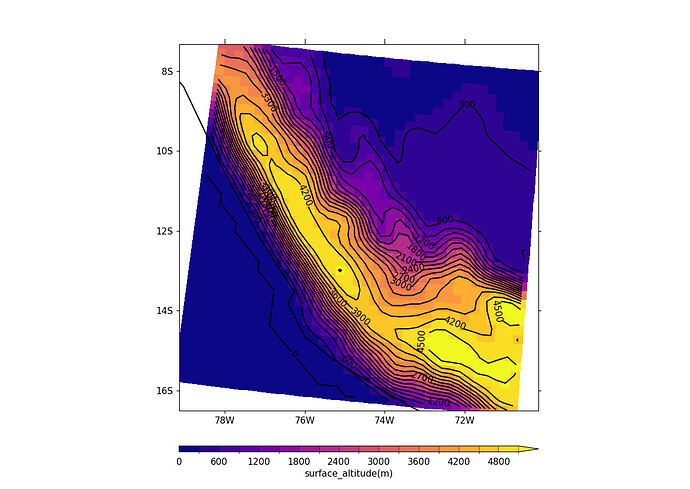
Hi Patrick,
The data projection is oblique mercator and isn’t in a format that Cartopy supports yet. Sadie’s done some detective work and it should be supported in the next release of cartopy. Sadie is also working on projections in cf-python so there will be two avenues to explore in the near future.
In the meantime we need to do a manual plot as cf-plot only accepts 1D lons and lats for coordinates when plotting blockfill.
import cf, cfplot as cfp, numpy as np
import cartopy.crs as ccrs
import matplotlib.patches as mpatches
g = cf.read('pmg2.nc')[0]
lons = g.coord('auxiliarycoordinate1').array
lats = g.coord('auxiliarycoordinate0').array
data = g.array
levs = [0, 300, 600, 900, 1200, 1500, 1800, 2100, 2400, 2700, 3000, 3300, 3600, 3900, 4200, 4500, 4800, 5100]
levs_arr = np.array([-100, 0, 300, 600, 900, 1200, 1500, 1800, 2100, 2400, 2700, 3000, 3300, 3600, 3900, 4200, 4500, 4800, 5100])
cfp.gopen()
cfp.levs(manual=levs, extend='both')
# Make a contour plot with alpha=0 so that the colour bar is plotted
cfp.con(g, alpha=0.0)
nx = np.shape(lons)[1]
ny = np.shape(lons)[0]
for ix in np.arange(nx - 1):
for iy in np.arange(ny - 1):
# Calculate the local size difference and set the square points
xdiff = (lons[iy, ix+1] - lons[iy, ix]) / 2
ydiff = (lats[iy+1, ix] - lats[iy, ix]) / 2
xpts = [lons[iy,ix]-xdiff, lons[iy,ix]+xdiff, lons[iy,ix]+xdiff, lons[iy,ix]-xdiff, lons[iy,ix]-xdiff]
ypts = [lats[iy,ix]-ydiff, lats[iy,ix]-ydiff, lats[iy,ix]+ydiff, lats[iy,ix]+ydiff, lats[iy,ix]-ydiff]
val = data[iy, ix]
try:
colour_index = np.max(np.where(val > levs_arr))
except:
colour_index = 0
# Plot the square
cfp.plotvars.mymap.add_patch(mpatches.Polygon(\
[[xpts[0], ypts[0]], [xpts[1],ypts[1]], [xpts[2], ypts[2]], [xpts[3],ypts[3]], [xpts[4], ypts[4]]],\
facecolor=cfp.plotvars.cs[colour_index], transform=ccrs.PlateCarree()))
cfp.gclose()
Here’s a plot using cfp.con(g) followed by the blockfill plot made using the code above
.